This study combines the CRISPR-Cas12a system with MRS sensing to create a CRISPR-Cas12a-based MRS biosensor for detecting Salmonella. The dual-functionality of MNPs as both magnetic separation tools and signal labels provides a new approach for biosensor design. Additionally, leveraging the high specificity of the CRISPR-Cas12a system and the high sensitivity of MRS detection, the biosensor exhibits excellent performance in detecting Salmonella.
A CRISPR-Cas12a-based MRS biosensor has been constructed to detect Salmonella. Small-sized MNPs were introduced as magnetic signal probes, and a new function for MNPs to generate signals was developed. The biosensor controls the binding of two different-sized MNPs using the CRISPR-Cas12a system when the target is present. It then utilizes the difference in magnetic separation speeds of these MNPs of different sizes to detect Salmonella using the small-sized MNPs as signal molecules. Thanks to the high specificity and efficient cutting activity of the CRISPR-Cas12a system, and the high sensitivity of MRS detection, the biosensor achieves LODs of 1.3×102 CFU mL-1 for pure bacterial liquid and 1.8×103 CFU mL-1 for spiked chicken meat extract samples. Additionally, it eliminates the need for repeated magnetic separations and washing steps, making the operation more convenient.
MNPs can serve as magnetic probes for MRS sensing detection. Traditional MRS biosensor methods based on MNP aggregation/disaggregation changes are susceptible to sample matrix interference and have poor stability. MRS biosensors based on MNP quantity changes have been well developed. However, these methods require the target itself to provide sites for controlling the binding between MNPs of different sizes. The limited surface sites of the target and possible non-specific binding may reduce the sensitivity of detection. Therefore, this paper combines the CRISPR-Cas12a system with MRS sensing technology, using CRISPR-Cas12a to precisely control the binding of two different-sized MNPs, and develops a CRISPR-Cas12a-based MRS biosensor for detecting Salmonella.
As shown in Figure 1, MNP130 and MNP30 were modified with SA and S2 on their surfaces to create the SA-modified MNP130 (MNP130-SA) and S2-modified MNP30 (MNP30-S2) probes. When the target is present, the Cas12a, crRNA, and target DNA form a ternary complex, activating the trans-cutting activity to non-specifically cut biotin-S1. As a result, MNP30-S2 cannot bind to the MNP130-SA surface through the base pairing between S1 and S2, and remains dispersed in the solution under the magnetic field. On the other hand, a large amount of MNP30-S2 binds to the MNP130-SA surface and is separated under the magnetic field. Finally, MNP30-S2 in the clear solution is used as a probe, and Salmonella is detected through the measurement of the transverse relaxation time T2.

Figure 1
Newmai NMR Support: The low-field NMR device (PQ001, Newmai Electronics Technology Co., Ltd., Shanghai, China) was used to measure the transverse relaxation time T2 of the samples. The instrument’s temperature was precisely set to 32±0.01°C. The instrument’s center frequency O1 and the 90o RF pulse width P1 and 180o pulse width P2 were calibrated using the hard-pulse sequence Q-FID. Then, the Carr-Purcell-Meiboom-Gill (CPMG) pulse sequence was used to measure T2.
1: TEM was used to characterize the surface state of MNP130-SA in the presence and absence of the target. As shown in Figure 2A, the particle size of MNP130-SA is around 130 nm, while the particle size of MNP30-S2 is around 30 nm. In the absence of the target, MNP130-SA captures biotin-S1 and binds with MNP30-S2, so a large amount of MNP30-S2 binds to the MNP130-SA surface. However, when the target is present, the trans-cutting activity of the Cas12a/crRNA complex is activated, non-specifically cutting biotin-S1, which affects the binding between MNP130-SA and MNP30-S2. As a result, only a small amount of MNP30-S2 is present around MNP130-SA. These TEM characterization results confirm the feasibility of the biosensor for detecting S. Typhimurium.
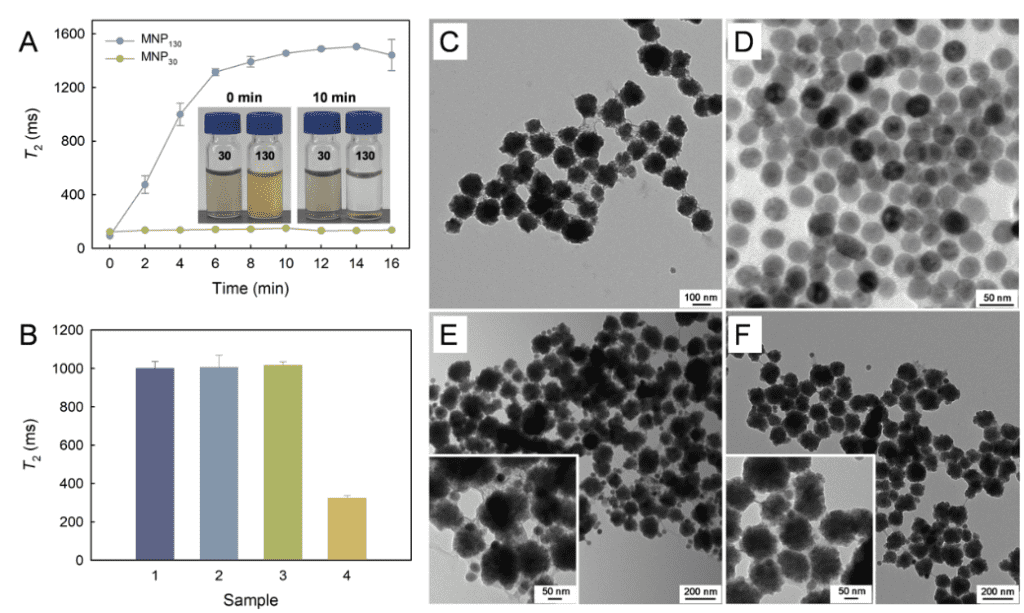
Figure 2
2: The effect of the cutting method on detection performance was evaluated. Scheme 1: biotin-S1 is first coupled to the MNP130-SA surface to form MNP130-S1, and then the CRISPR-Cas12a system cuts the S1 on the MNP130 surface. Scheme 2: the biotin-S1 in the solution is cut first, then MNP130-SA is added for capture. Using Scheme 1, the target can be qualitatively detected by observing changes in dispersion, which is simple and convenient but has lower sensitivity. However, using Scheme 2 significantly improves the MRS biosensor’s response to the target. To improve detection sensitivity, Scheme 2 will be used in subsequent experiments for detecting Salmonella.
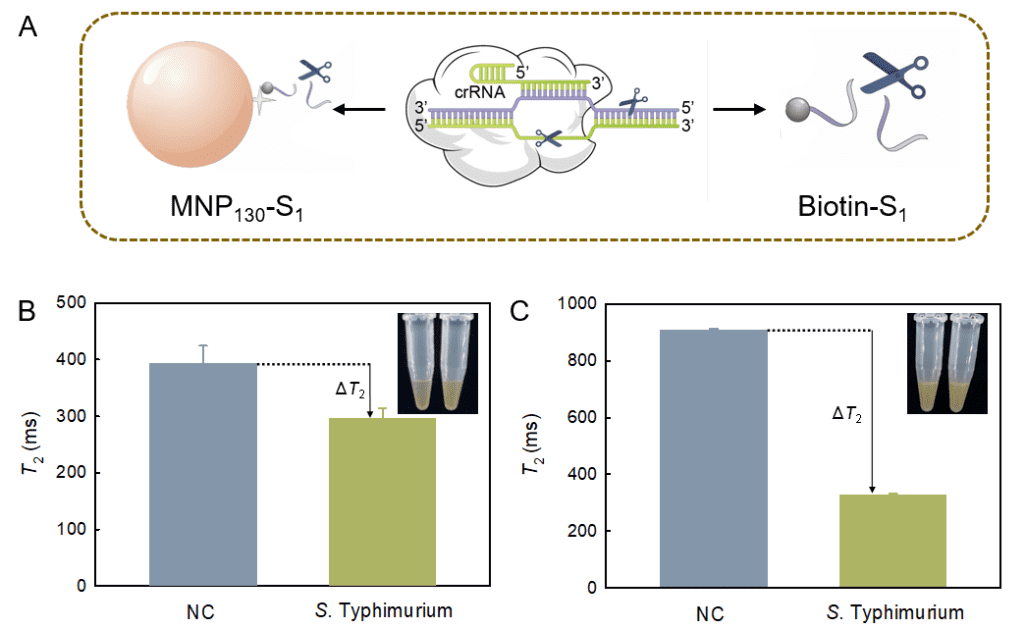
Figure 3
3: From Figure 4, it can be seen that at a bacterial concentration of 1.3×102 CFU mL-1, the T2 value is below the threshold line, indicating that this method can detect S. Typhimurium at 1.3×102 CFU mL-1, with an LOD significantly lower than most reported MRS biosensors. Detection of S. Typhimurium in spiked chicken meat extract samples yields an LOD of 1.8×103 CFU mL-1. A comparison of the detection results between pure bacterial liquid and chicken meat extract samples shows that the signal response caused by bacteria at 103 CFU mL-1 in the chicken meat extract is significantly higher than that in pure bacterial liquid (ΔT2% of 10% and 47%, respectively). To evaluate the specificity of the biosensor for Salmonella, four common foodborne pathogens, S. aureus, E. coli O157:H7, V. parahaemolyticus, and L. monocytogenes, were selected for investigation. S. Typhimurium and the four non-target bacteria all had concentrations of 105 CFU mL-1. As shown, compared to the blank control, the signal change rates caused by non-target bacteria were all within 5%, while S. Typhimurium caused more than a 60% signal reduction, demonstrating the excellent specificity of the biosensor.
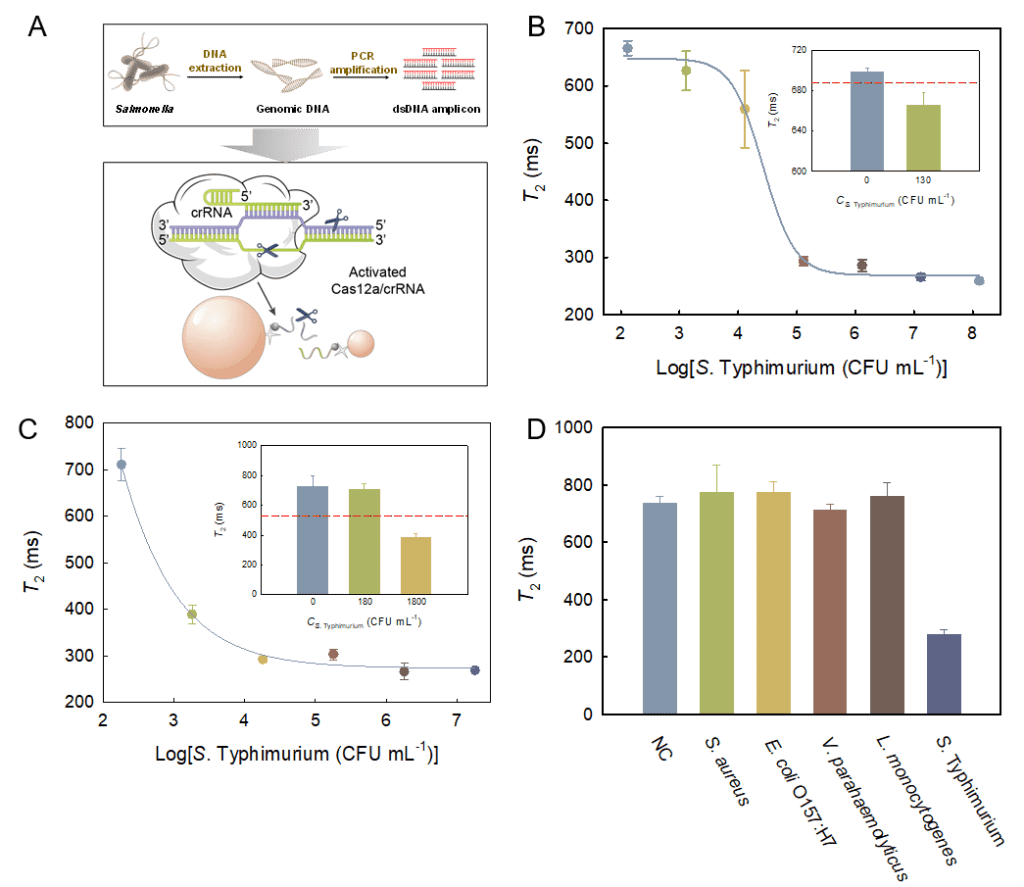
Figure 4
This study presents a CRISPR-Cas12a-based MRS biosensor for detecting Salmonella. The method uses the CRISPR-Cas12a system to control the binding of two different-sized MNPs, with free small-sized MNPs serving as magnetic probes for MRS signal output. The LODs for S. Typhimurium in pure bacterial liquid and spiked chicken meat extract samples are 1.3×102 CFU mL-1 and 1.8×103 CFU mL-1, respectively. This method takes full advantage of MNPs’ dual function as both magnetic separation carriers and signal labels. Furthermore, by measuring the clear solution after magnetic separation, it eliminates tedious repeated washing steps, simplifying the detection process. The method can be extended to the detection of other targets by designing specific crRNA, demonstrating its general applicability.
This research was supported by the National Natural Science Foundation for Young Scholars, the Walmart Foundation, and the Walmart Food Safety Collaboration Center.

Phone: 400-060-3233
After-sales: 400-060-3233


Back to Top